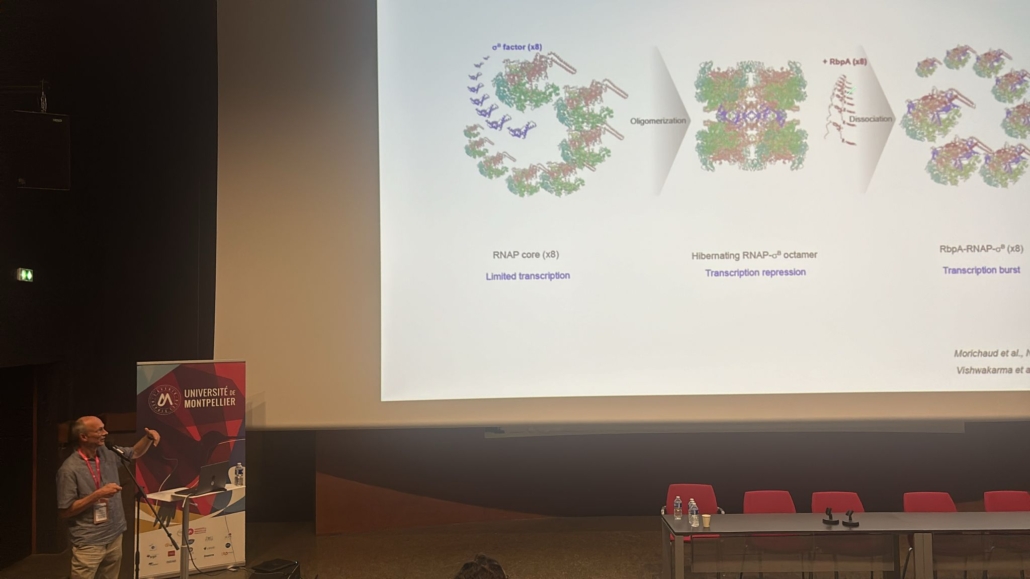
Axe 1: l’ARN polymerase bactérienne et la résistance aux antibiotiques (K. Brodolin)
L’ADN-dépendante polymerase de l’ARN (ARNP) est l’enzyme centrale de l’expression génique et une cible pour la régulation génétique. Ses caractéristiques de base sont hautement conservées parmi tous les organismes vivants. l’ARNP bactérienne est une cible pour un grand nombre de protéines de régulation et de petites molécules, notamment des antibiotiques. Parmi les molécules antibactériennes inhibant l’ARNP, la rifampicine (Rif) reste un antibiotique de première ligne pour le traitement de la tuberculose. Récemment, une nouvelle classe d’antibiotiques: lipiarmycin (fidaxomicine, Tiacumicin B) et myxopyronin (Myx) a été caractérisée. Le mécanisme d’action de ces molécules reste à élucider. Les bactéries ont développé des mécanismes multiples pour échapper aux effets du traitement antibiotique. La plupart de ces mécanismes, tels que l’activation des voies de réponse au stress et le passage à l’état persistant sont liées à la régulation de l’activité de l’ARNP. Une quantité croissante de preuves indique que la sous-unité sigma de l’ARNP ainsi que d’autres facteurs transcriptionnels (par exemple la RbpA de M. tuberculosis) sont impliqués dans la génération de la résistance en réponse au traitement antibiotique. Les objectifs globaux de nos études sont (1) comprendre la rôle de l’interaction entre la sous-unité sigma et les facteurs transcriptionnels dans la modulation de l’expression génique; (2) déchiffrer les mécanismes d’action des antibiotiques ciblant l’ARNP (3) comprendre comment les bactéries résistent à ces molécules. Notre projet est axé sur le mécanisme de régulation de la transcription de deux agents pathogènes humains: Escherichia coli, qui est considéré comme un prototype pour les bactéries pathogènes, et Mycobacterium tuberculosis qui est d’une grande importance clinique. Nous explorons également le rôle de la sous-unité sigma dans les étapes clés de l’expression des gènes tels que la reconnaissance de promoteur, la synthèse de l’ARN et pause de la transcription. Les résultats de notre étude contribuent à la compréhension de la base moléculaire de la résistance aux antibiotiques, constituent une base pour le développement de nouveaux médicaments, plus efficaces en pharmacologie.
Axe 2: Développement de nouvelles stratégies thérapeutiques anti-infectieuses (L. Chaloin)
L’objectif de nos études est de développer de nouvelles armes thérapeutiques pour mieux lutter contre certaines infections virales et bactériennes. Une thématique en lien avec la précédente consiste à mieux comprendre l’action des antibiotiques dirigés contre l’ARN polymérase avec notamment le repositionnement de la Fidaxomicine (initialement développée contre Clostridium difficile) pour inhiber la transcription chez M. tuberculosis. Des études biochimiques, structurales et théoriques sont menées en parallèle afin d’élucider les mécanismes d’action, le développement de résistance, la mise au point de nouveaux antibiotiques.
Deux autres axes thématiques sont développés en collaboration avec l’équipe APIR (J.-M. Péloponèse) pour lutter contre les virus à ARN (HIV-1, HTLV-1, Chikungunya, Dengue et SARS-CoV-2) en ciblant l’ARN hélicase A qui est utilisé et nécessaire à la multiplication de ces virus. En parallèle une seconde approche est dédiée à enrayer la transformation des cellules infectées par HTLV-1 conduisant à la leucémie T de l’adulte. La stratégie appliquée consiste à développer des inhibiteurs spécifiques de la protéine virale HBZ (HTLV-1 bZip factor), cette dernière protéine codée par le brin antisens du génome viral est exprimée tout au long du processus d’infection mais également après la transformation au stade leucémique, ce qui en fait une cible privilégiée.
Pour ces axes thématiques à visées thérapeutiques, des méthodologies de criblage virtuel, simulation de dynamique moléculaire sont utilisée en parallèles des données expérimentales obtenues au sein de l’équipe.
POST-DOC, THÈSES, STAGES
Nous accueillons des candidats (biologistes, biochimistes, biophysiciens, physico-chimistes, physiciens) mais aussi des chercheurs et des ingénieurs et techniciens en poste (CNRS, INSERM, Université) intéressés par des approches interdisciplinaires de problèmes biologiques à l’échelle de la molécule unique ou d’assemblages moléculaires..
En bref
Organisme(s) modèle(s)
Escherichia coli, Mycobacterium tuberculosis
Processus biologique étudié
Transcription, Résistance aux antibiotiques
Techniques utilisées
- Biochimie
- Biophysique structurale (cristallographie, microscopie électronique)
- Bactériologie
- Modélisation moléculaire
Applications médicales
- Développement de molécules anti-infectieuses
Financements