Our team is interested in the role of viral and cellular proteins and their interactions with viral RNA and host cell lipids during viral translation and virus assembly. We study these phenomena quantitatively at the single-molecule level in model systems, in living host cells and multi-cellular organisms.
Our research focuses on 6 main topics
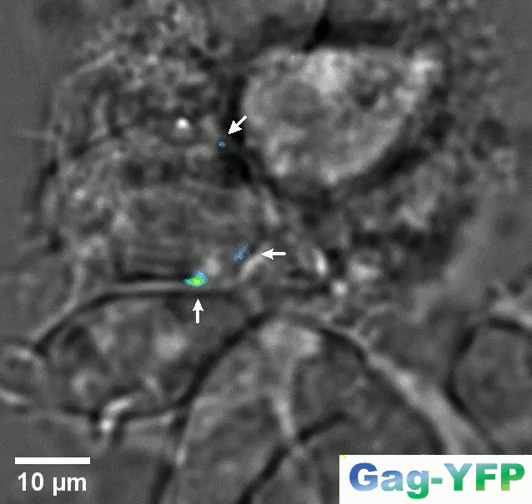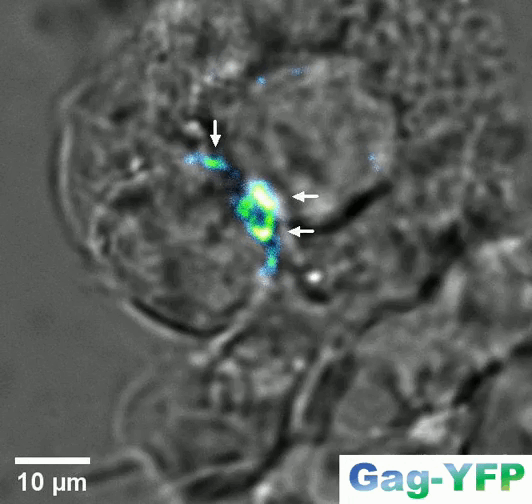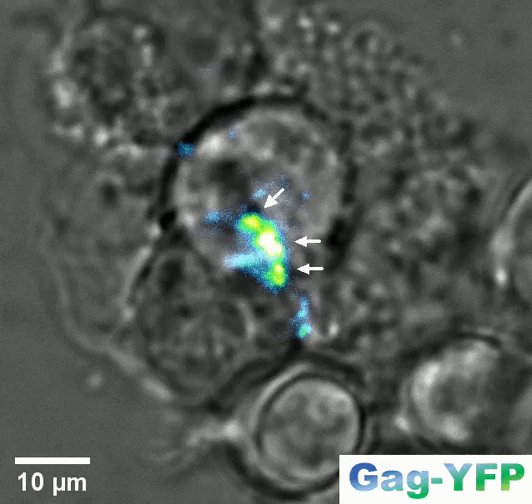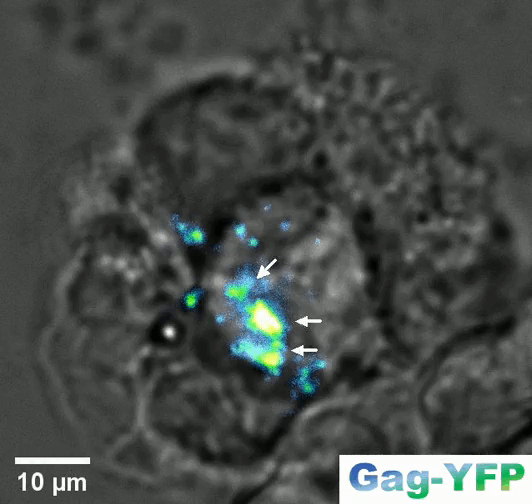
Retroviral biofilm assembly and polarization at the T cell-T cell junction (from Arone et al., mBio 2023)
- Lipid sorting during the self-assembly of HIV-1 Gag and influenza A M1 proteins.(Kerviel 2013; Saad & Muriaux, 2015; Kerviel 2016; Yandrapalli 2015, 2016; Favard 2019)
- Single molecule microscopy of HIV-1 assembly in live CD4 T cells. (Mariani 2016; Mariani 2018; Inamdar 2019; Dibsy 2023)
- Role of membrane co-factors and cortical actin during viral assembly. (Thomas 2015; Inamdar 2021; Dibsy 2023)
- Role of M-matrix proteins and cellular cofactors in influenza virus, RSV and SARS-CoV-2 assembly. (Kerviel 2016; Bracquemond & Muriaux, 2021; Gourdelier 2023; Swain, 2023; Swain, 2023)
- Atomic force microscopy (AFM) study of HIV-1, IAV and SARS-CoV-2 assembly on single virus particles. (Faivre-Moskalenko 2014; Bernaud 2015; Lyonnais 2021)
- Dynamic of viral replication (translation/amplification) in cells and developing organisms. (Dufourt et al., Science 2022; Bellec et al., Nat Comm 2022; Bellec et al., RNA 2024)
1. Lipid sorting during self-assembly of HIV-1 Gag and influenza A M1 proteins
In previous studies, we have shown that the interaction between the Matrix domain of the Gag protein and specific plasma membrane lipids (PS/PIP2) induces retrovirus assembly (Hamard-Péron et al., 2010 ; Hamard-Péron & Muriaux, 2011). We proposed that acidic lipid-enriched microdomains (ALEMs) are created by oligomerization of the HIV-1 Gag protein in the host cell plasma membrane during viral assembly (Kerviel et al., 2013, Yandapalli et al., 2014, Mariani et al., 2014 & Saad & Muriaux, Editorial 2015). We first have characterized the nature and the effects of the MA domain interaction of retroviral Gag proteins (HIV-1 and MLV) with lipid membranes in silico (Kerviel et al., 2013, Charlier et al., 2014). In vitro, we investigated the impact of HIV-1 Gag protein self-assembly on lipid plasma membrane using model membranes with controlled molecular composition mimicking this membrane. We were able to show that, during its self-assembly, the viral Gag protein sorts membrane lipids and generates lipid nano-domains enriched in PIP2 and Cholesterol, while excluding Sphingomyelin (Yandrapalli et al., 2016).
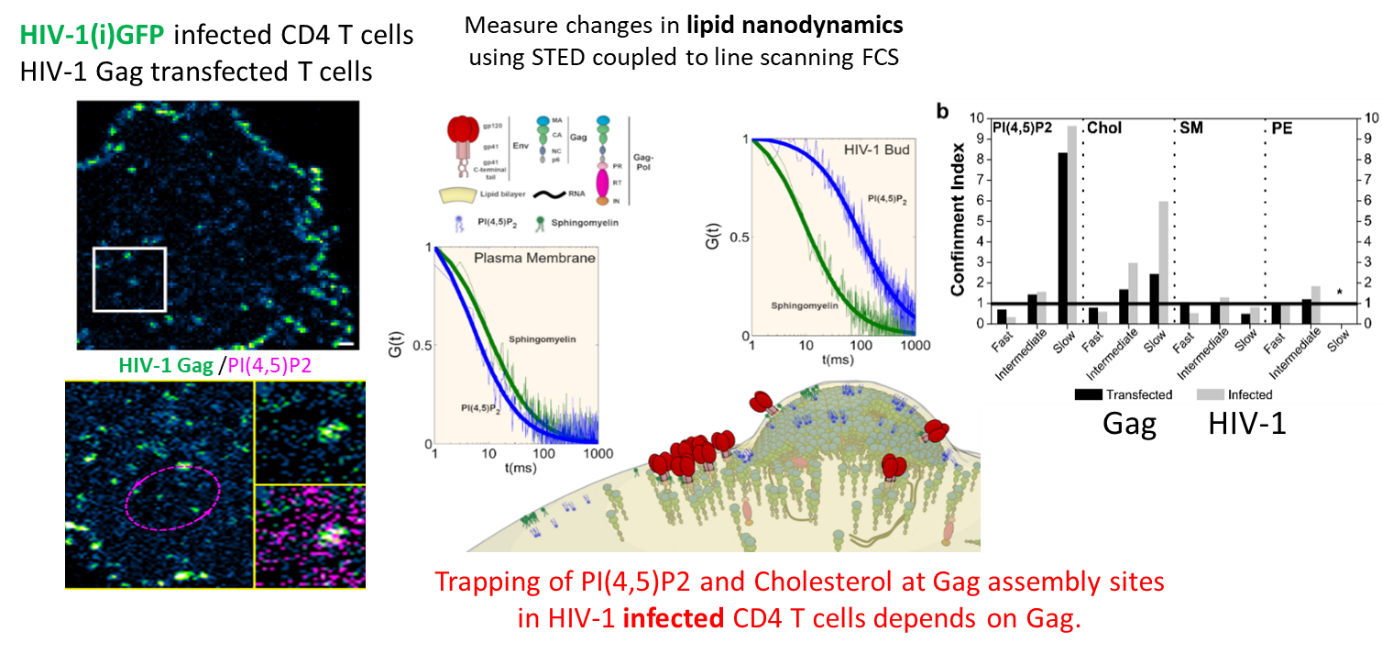
We finally quantified this phenomenon in viral host cells (HIV-1-infected or Gag-transfected CD4 T cells) by measuring the diffusional changes of lipids (PIP2, Cholesterol, SM and PE) into and out of the viral bud on the T cell surface by sSTED-FCS (Scanning STED FCS), in collaboration with C. Eggeling (U. Oxford/ Univ Jena, UK-Germany) and J Chojnacki (U. Oxford/ IRSI Caixa, Barcelona, SP) (Favard et al., 2019).
This work was highlighted by the Company of Biologists, by the CNRS-INSB in its monthly news as well as in its 2019 highlights and also by ANRS.
2. Single molecule microscopy of HIV-1 assembly in living CD4 T cells
In parallel, we have studied the dynamics of capsid protein assembly in the cell using super-resolution optical microscopy methods (Live PALM, spt-PALM), in collaboration with the teams of JB Sibarita (IINS, Bordeaux) and M. Dahan (Institut Curie, Paris).
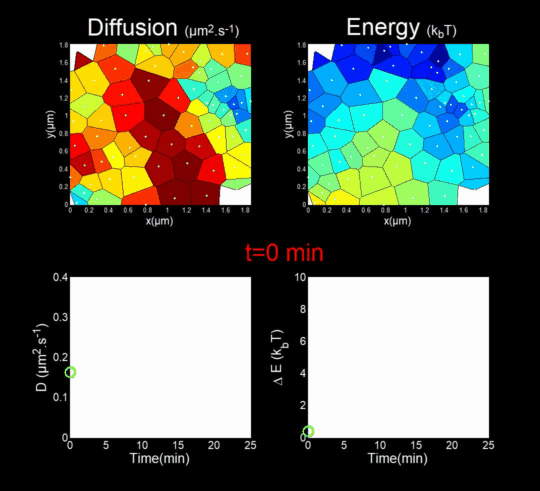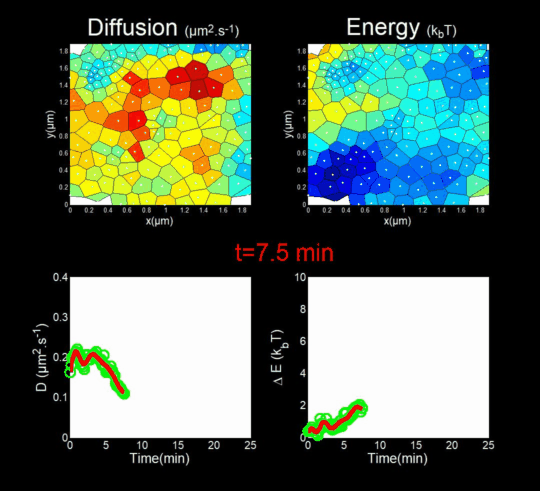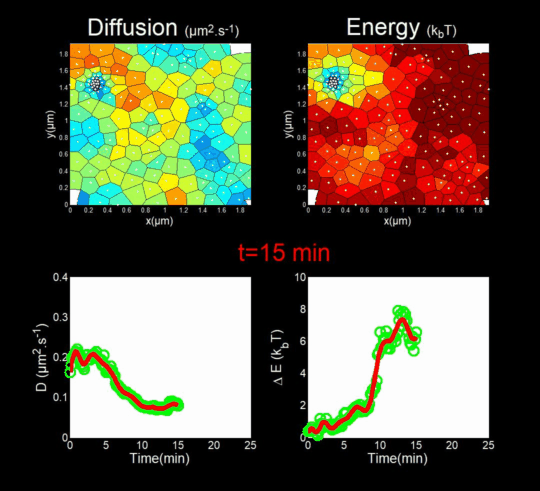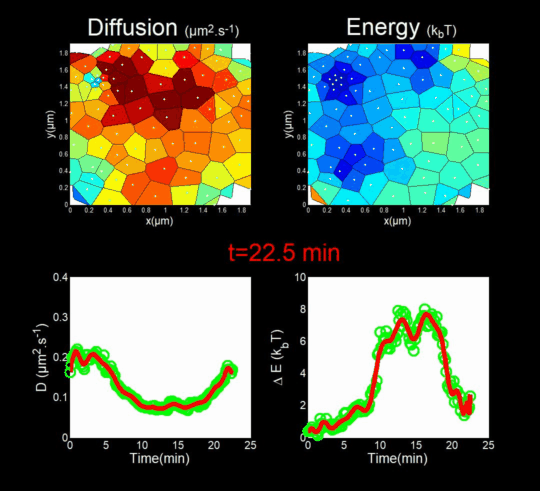 We followed the Gag self-assembly, molecule by molecule, at a resolution of a tenths of nanometers, generating billions of individual trajectories. (Mariani et al., 2016, Journal Cover)
We followed the Gag self-assembly, molecule by molecule, at a resolution of a tenths of nanometers, generating billions of individual trajectories. (Mariani et al., 2016, Journal Cover)
We rely on methods from Big Data, for the automatic analysis of these trajectories using Bayesian inferences. In collaboration with J.B. Masson (Institut Pasteur), M. El Beheiry and M. Dahan (I. Curie), we were able to follow, during 25 minutes, molecules after molecules, the self-assembly of Gag during the formation of a VLP of HIV-1 in a T lymphocyte. This was done by measuring both the changes in the diffusion of these molecules (left part of the movie) but also in the interaction energy (right part of the movie) that is sensed by each Gag in the vicinity of the assembly site. (Mariani et al., 2018)
Importantly, we found the Gag-Gag interaction energy in living T-cells to be similar to the one predicted by molecular dynamics.
Our work on the study of HIV-1 assembly using super-resolution microscopies of the living has resulted in 2 general interest reviews (Inamdar et al., 2019 et Arone et al., 2021).
3. Role of membrane co-factors and cortical actin during viral assembly
This project aims to understand the role of filamentous cortical actin (F-actin) and newly identified cellular co-factors from HIV-1 host cells (T-CD4+ lymphocytes) in the modulation of the dynamics and organization of membrane domains as well as in the membrane curvature generation during HIV-1 assembly as well as filamentous cortical actin (F-actin).
In our previous research, we have shown that CD81 tetraspanin is a host cell transmembrane protein involved in HIV-1 assembly in CD4 (Grigorov et al., 2009) and that the clathrin-dynamin endocytosis pathway was involved in the transmission of HIV-1 from CD4 T cells to CD4 T cells (Bosch et al., 2008). We are now interested in studying the role of these tetraspanins in the assembly of another human retrovirus HTLV-1 chronically infecting T cells in collaboration with Hélène Dutartre (CIRI,ENS Lyon) (Arone et al., 2023)
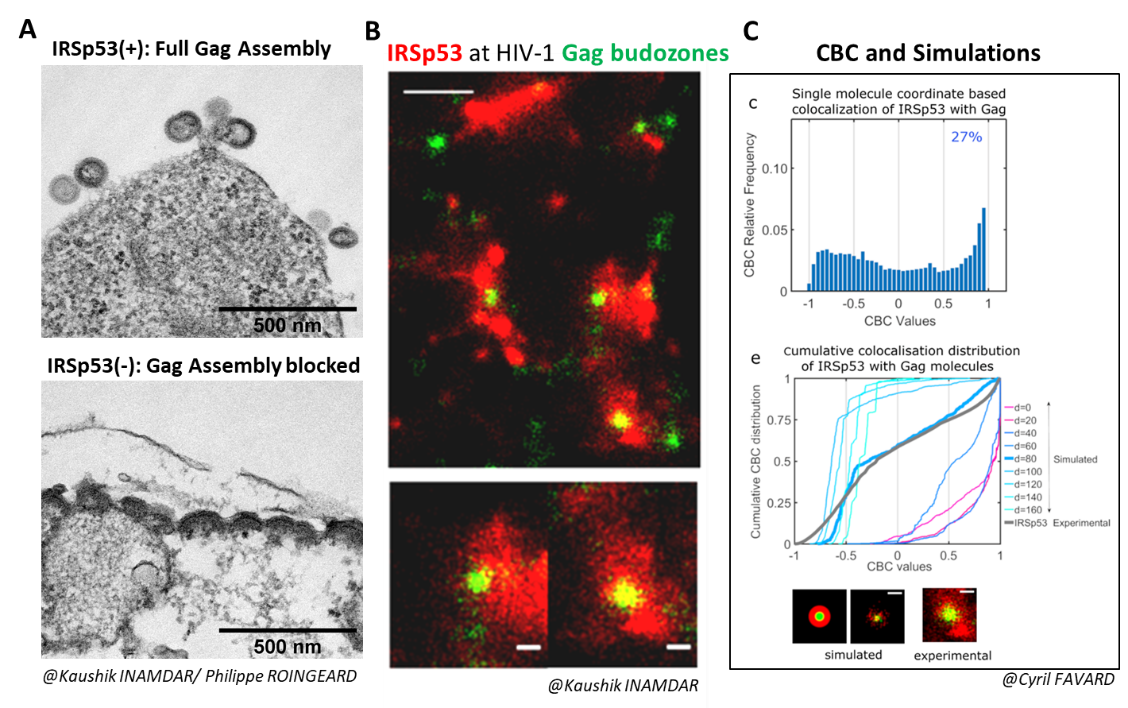
From Inamdar et al., 2021.
In parallel, we also studied the role of a major regulator of the cortical actin network regulated by the Rac1/IRSp53/Wave2/arp2/3 activation pathway in the assembly of the HIV-1 Gag protein and the release of the viral particle in CD4 T cells, the main hosts of HIV-1. (Thomas et al., 2015).
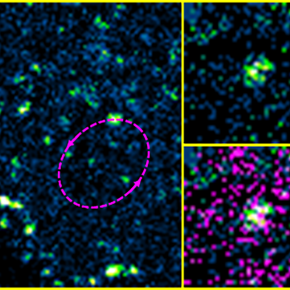
We recently showed that the membrane bending factor IRSp53 was indeed involved in the assembly and budding of the HIV-1 Gag viral particle (Inamdar et al., 2021). During the reduction of IRSp53 expression by interfering RNA in HIV-1 Gag particle-producing cells, we observe a blockage of virus assembly at midstream (Figure A). IRSp53 is found at the site of virus assembly, as observed by PALM (Gag)/STORM (IRSp53) microscopy (Figure B) and quantified by correlating experimental single molecule coordinate based colocalization (CBC) measurements with those obtained by numerical simulations on pre-defined models (Figure C). The cellular protein IRSp53 assists in membrane bending during HIV-1 particle formation to finalize HIV-1 assembly and budding.
4. Role of M-matrix proteins and cellular cofactors in the assembly of influenza virus and SARS-CoV-2
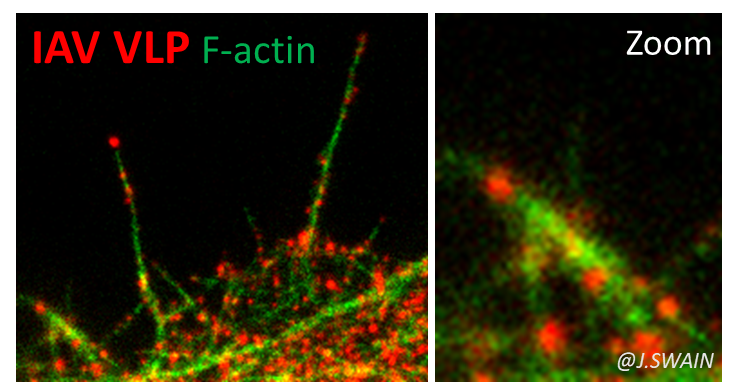
We are currently studying other enveloped RNA viruses. The influenza virus, A (H1N1) pdm09, still circulating, which caused the first pandemic of the 21st century. We are interested in the role of the matrix protein M1 (and M2) in the membrane assembly of this virus. M1 is the most abundant viral protein and is essential for influenza virus assembly. We have recently shown that the arginine triplet (R76/77/78) located on helix 5 of M1, a highly conserved motif among influenza A subtypes, is essential for its localization to the cell plasma membrane, for the membrane attachment of M1, for its incorporation into viral particles and for its infectivity (in collaboration with O.Moncorgé, IRIM CNRS Montpellier and P. Roingeard, University of Tours). Its mutation completely abolishes the production of infectious virus. This study also allowed us to develop a minimal system for the production of non-infectious influenza A VLP-M (Kerviel, Dash et al., 2016).
We are now interested in the role of cortical actin and its dynamic regulators in the assembly and budding of influenza A virus and SARS-CoV-2 (Bracquemond & Muriaux, 2021), that we explore using RNA interference, infection, and cell biology techniques coupled with advanced photonic microscopies (Swain et al., 2023).
5. HIV-1, IAV and SARS-CoV-2 assembly visualized by Atomic force microscopy (AFM) on single virus particles
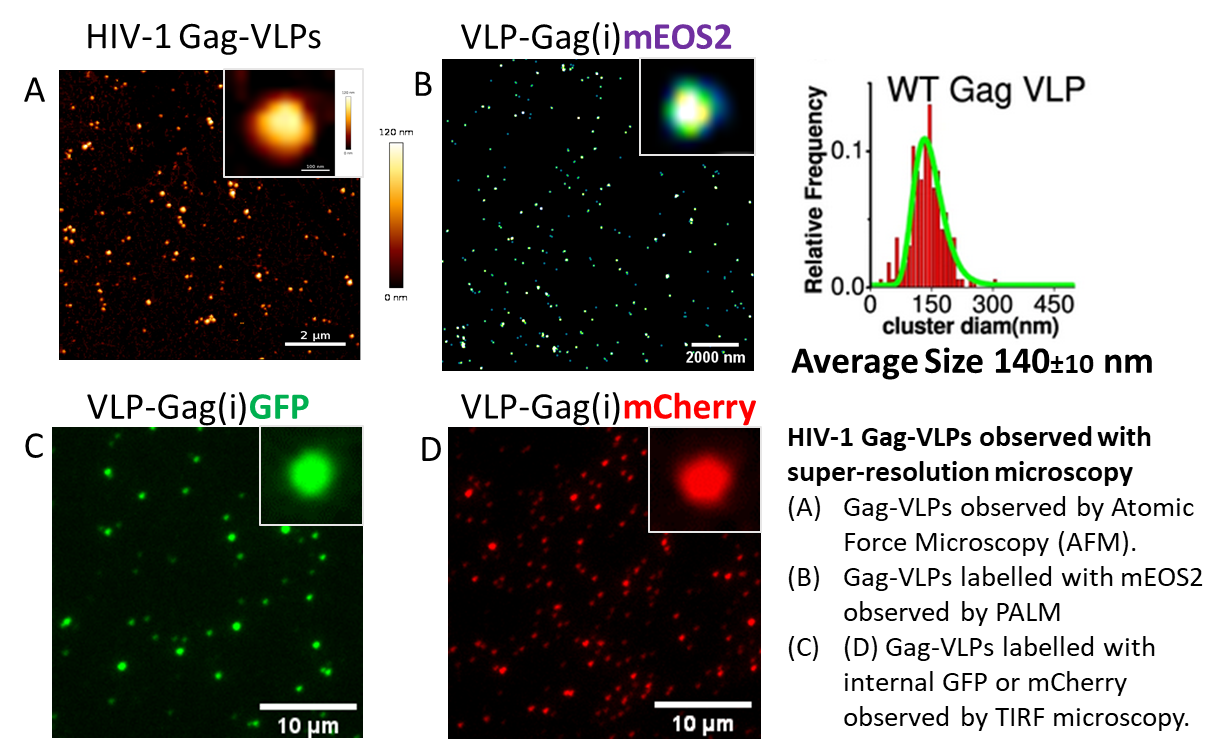
We have shown using atomic force microscopy at the level of single virus particles that genomic RNA is a polydispersity factor in the morphology of HIV-1 (Faivre-Moskalenko et al.,2014). In collaboration with the physicists C. Moskalenko and M. Castelnovo at ENS Lyon, atomic force microscopy is used to quantitatively study the variation of the size of viral particles and cores produced by host cells, as a function of various micro-environments (Bernaud et al., 2015). This polydispersity is an estimator of the self-assembly efficiency of the HIV-1 Gag protein.
We have recently imaged the infectious and the inactivated SARS-CoV2 virus by AFM in collaboration with Sébastien Lyon and the Cemipai team (Lyonnais et al., 2021).
We continue to analyze all types of VLPs derived from HIV-1, IAV and SARS-CoV-2 viruses by these fluorescence techniques, which represent wonderful tools to study the assembly or the entry of viruses into their host cell (outside a BSL3).
6. Dynamic of viral replication (translation/amplification) in cells and developing organisms.

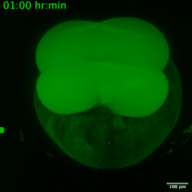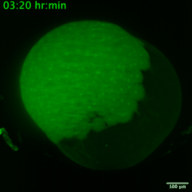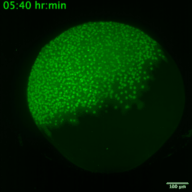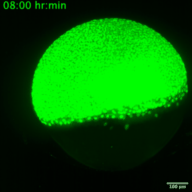
RNA viruses represent a major public health problem worldwide. Some of these RNA viruses are implicated in emerging infectious diseases. As humanity recovers slowly from the COVID-19 pandemic, the expansion of arthropod vector-borne viruses (arboviruses) requires an immediate awakening to avoid a new health world crisis. Arboviruses mostly belong to two families of positive sense single-stranded RNA viruses: Flaviviridae and Togaviridae including viruses such as Zika, Chikungunya and Sindbis. In humans, the most common clinical symptoms of infection are fever, headache, and pain, but they can reach the nervous system and lead to encephalitis and neuronal disorders. Fundamental research into their replication cycles and the associated immune response is necessary, and would facilitate the development of new treatments. Indeed, viral genome localization, translation and replication are complex and highly regulated processes that occur during the replication of RNA viruses. These processes are tightly orchestrated, and their synchronization is crucial for the formation of new viruses. Although RNA viruses have been widely studied, little is known about the spatio-temporal processes that take place during viral infection in living organisms (e.g., what are the dynamics of viral translation? at which time scale are host cells able to respond to infection?). Moreover, these dynamics can vary according to cell type and developmental stage at the time of infection. Thanks to the recent development of methodologies enabling us to study the live translation of single mRNAs in a living organism, we propose to dissect, in vivo, the interaction between the translation/replication dynamics of Zika, Chikungunya and Sindbis viruses and the immune response of infected cells. We will use the zebrafish as a model organism to study viral cycle dynamics and the associated immune response during development, with high spatio-temporal resolution and at the single-molecule level.
Team Leaders

We are studying the mechanisms involved in the assembly of enveloped viruses, from single molecules to organoids.
We are biologists and biophysicists using multi-disciplinary approaches and cutting-edge optical microscopies.
Financial Supports