
The AMI team studies the interactions between pathogens (bacteria and viruses) and the autophagy machinery, including canonical autophagy and Atg8ylation, throughout pathogen life-cycle, from entry into the target cells to replication and cellular homeostasis. This project, which relies on fundamental aspects of the interactions between autophagy-related processes and pathogens, will lead to a better understanding of the mechanisms of infections.

The AMI team in May 2025, from left to right: Nathalie Chazal, Guilhem Cantaloube, Lucas Boulet, Romane Deteve, Sébastien Lainé, Marie Villares, Jerôme Feuillard, Julie Couston, Fiona Nicole, Mickaël Blaise, Coralie Daussy, Célia Chamontin, Marylène Mougel, Lucile Espert.
Our team studies the interactions between pathogens (viruses and bacteria) and the host cell response throughout the pathogen’s life cycle. In particular, we investigate the role of autophagy mechanisms and, more broadly, atg8ylation, during infections. Our projects are based on fundamental aspects that aim to better understand the mechanisms involved in infections, with the goal of identifying targets for the development of translational approaches that could lead to new anti-infectious strategies.
Our projects are structured around two main axes:
1. Role of autophagy and atg8ylation in the entry of pathogens into their target cells.
Pathogen entry is, by definition, the first event in the microbial life cycle that triggers a cellular response. This response can radically alter the outcome of the infection because, during and shortly after entry, pathogens have not yet expressed their full genetic repertoire, giving the cell a significant selective advantage to block the infection. The aim of this research axis is to determine the mechanisms by which the autophagy machinery is involved in the entry of pathogens into their target cells. Our work has notably shown that atg8ylation promotes HIV-1 entry via membrane fusion in CD4+ T lymphocytes, independently of autophagy. We are investigating whether this process is common to the entry of other enveloped viruses, particularly SARS-CoV-2. We are also exploring the role of autophagy and atg8ylation in the early stages of infection of macrophages by different Nocardia species, for which nothing is currently known.
2. Role of autophagy and atg8ylation in pathogen replication and the maintenance of cellular homeostasis.
The autophagy pathway is well known for controlling infections and maintaining cellular homeostasis. Pathogens exploit or neutralize these mechanisms to ensure their replication, potentially impacting the survival of infected cells. In this context, we are studying the role of autophagy in the innate immune response triggered by infection with various enveloped viruses. The role of the autophagy machinery in maintaining cellular homeostasis during infections is also being investigated. In the case of the Nocardia bacterial model, nothing is currently known about its replication mechanisms within host cells. However, the fact that this bacterium multiplies efficiently in its target cells strongly suggests that it has adapted to block or use the autophagy machinery to its advantage. We will therefore use various approaches to determine the bacterial determinants that allow Nocardia spp to survive and multiply, by investigating their interactions with the autophagy machinery.
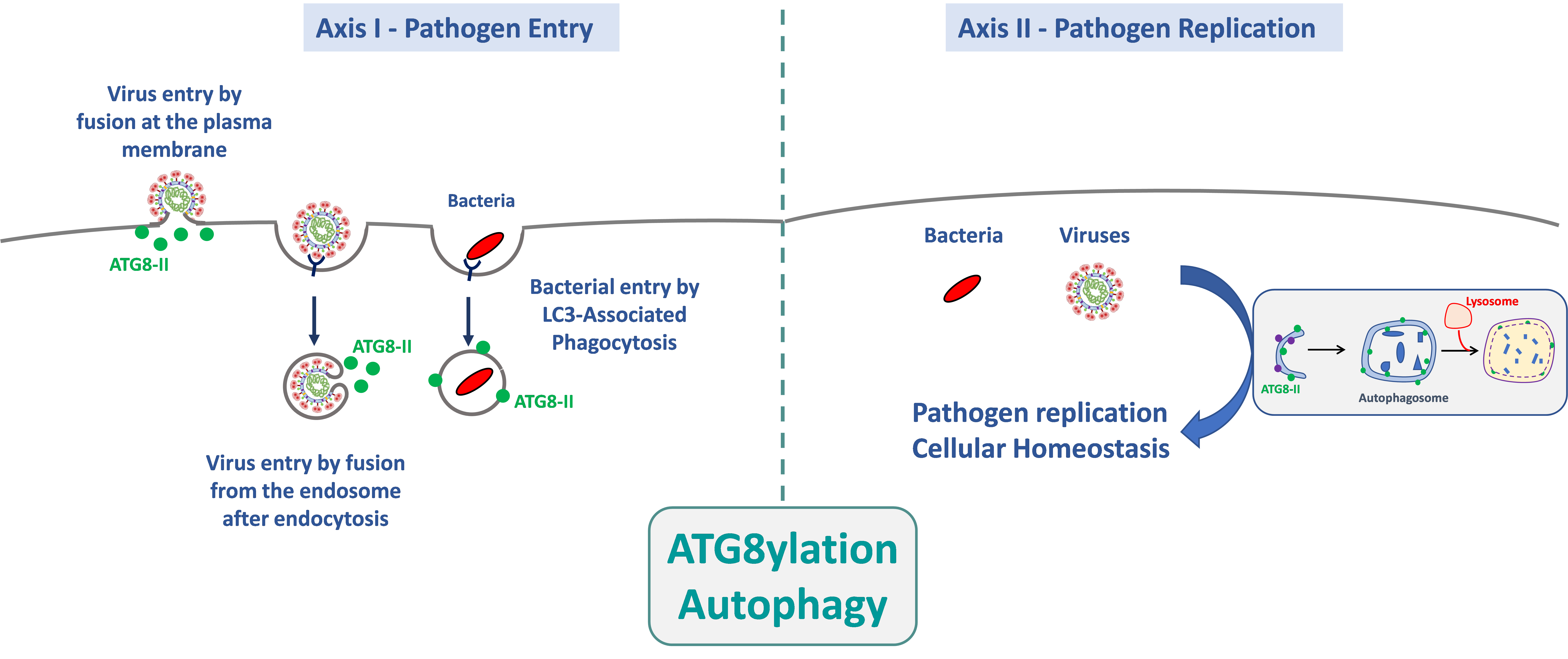
The AMI team studies the interactions between pathogens (bacteria and viruses) and the autophagy mechanism, and more broadly atg8ylation, throughout the pathogen’s life cycle—from its entry into target cells to its replication and its role on cellular homeostasis. Our project, which is based on fundamental aspects of the interactions between autophagy-related processes and pathogens, will provide a better understanding of the cellular mechanisms involved during infections.
3. Identify drug-resistance mechanisms to first-line treatment and to beta-lactams and aminoglycosides.
Nocardia spp. are considered emerging pathogens as the number of infections is expected to increase in parallel to chronic affections, such as chronic obstructive pulmonary disease (COPD), in coming years. Nocardiosis can be treated by first-line treatment made of two drugs trimethoprim and sulfamethoxazole targeting the dihydrofolate reductase and the dihydropteroate synthase. These two enzymes are involved in the synthesis of folate. Since the treatment lasts as long as several weeks to months drug resistance to these two drugs is often reported. For severe infections, the first-line drugs are often associated with beta-lactams or aminoglycosides. Antimicrobial resistance (AMR) to both drugs has been however reported but is not well-understood. Thus, understanding these AMR can help combat Nocardia species infections.
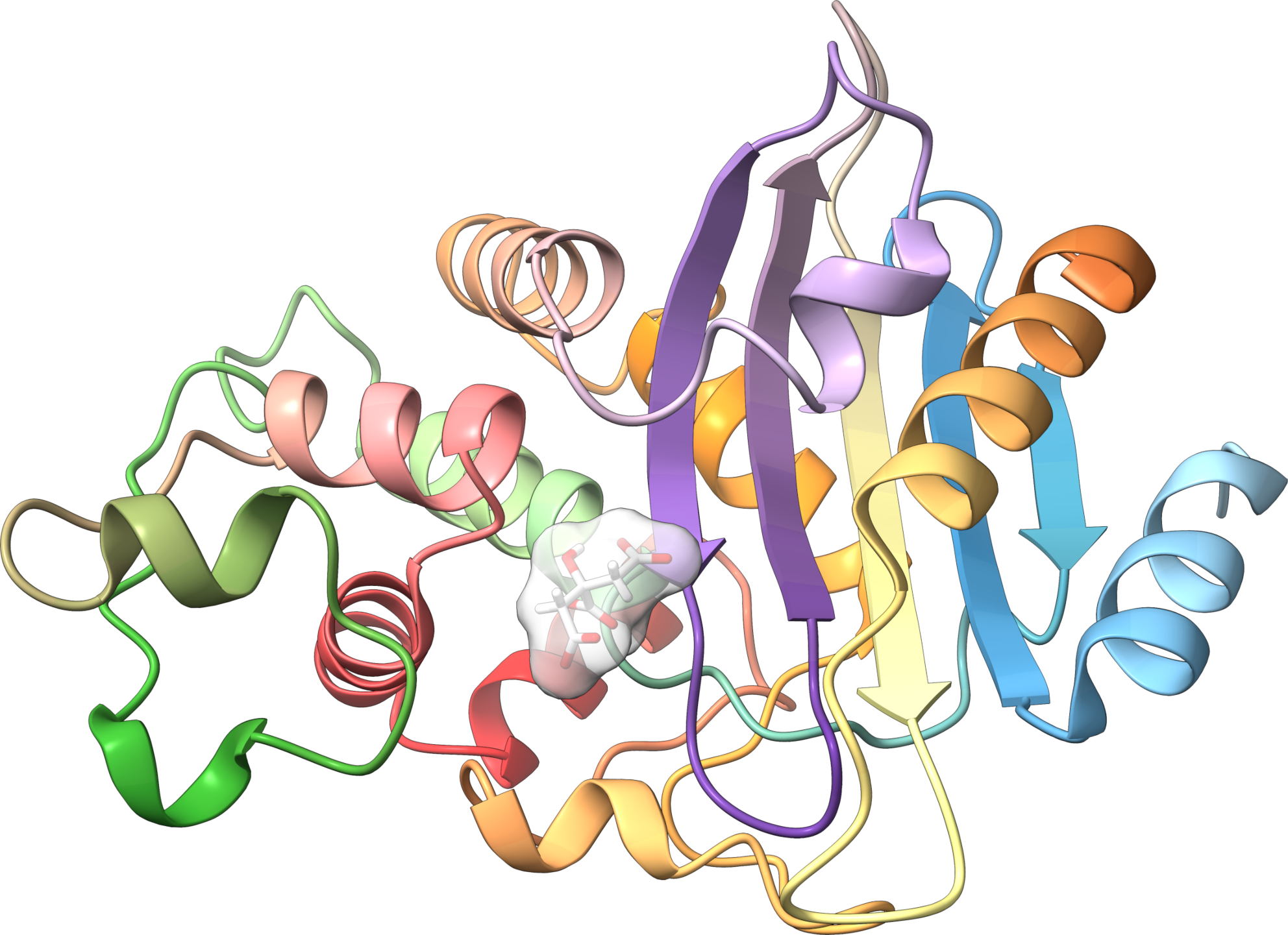
Crystal structure of a class A beta-lactamase from N. cyriacigeorgica that confers resistance to penicillin derivatives
Key words: Autophagy – Atg8ylation – Enveloped viruses – Nocardia spp – Antimicrobial resistance – Structural biology









