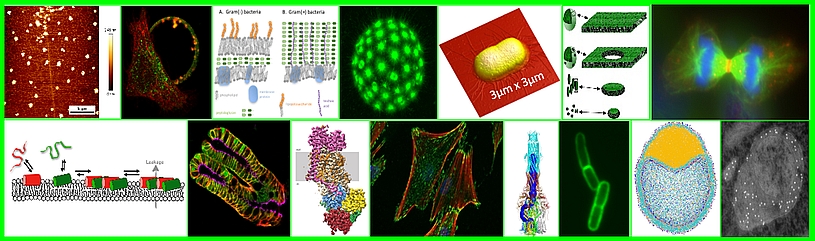
Our team is interested in the role of viral and cellular proteins and their interactions with viral RNA and host cell lipids during viral translation and in virus assembly. We study these phenomena quantitatively at the single-molecule scale in model systems, in host cells and living multicellular organisms.
Our research focuses on six major questions:
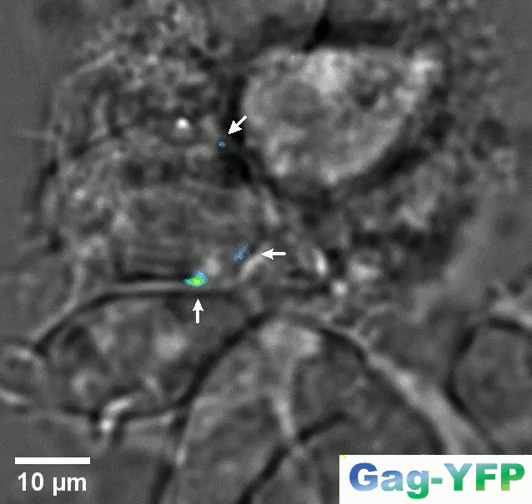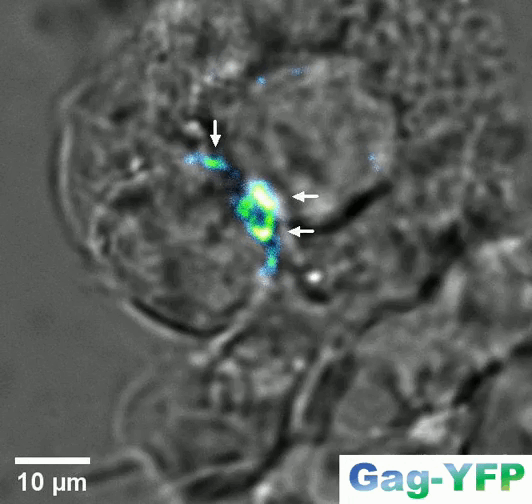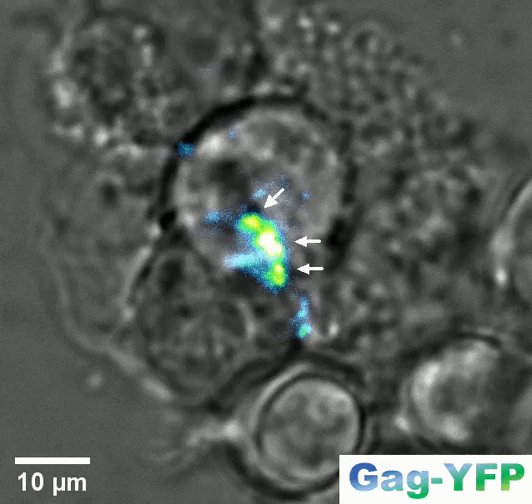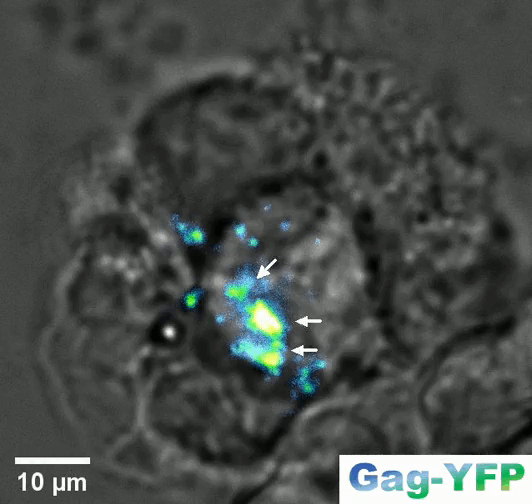
Retroviral biofilm assembly and polarization at the T cell–T cell junction (from Arone et al., mBio 2023)
- Lipid sorting during the self-assembly of HIV-1 Gag and influenza A M1 proteins.
(Kerviel 2013; Saad & Muriaux, 2015; Kerviel 2016; Yandrapalli 2015, 2016; Favard 2019)
- Single-molecule microscopy of HIV-1 assembly in living CD4 T lymphocytes.
(Mariani 2016; Mariani 2018; Inamdar 2019; Dibsy 2023)
- Cortical actin dynamics and membrane curvature during viral assembly.
(Thomas 2015; Inamdar 2021; Dibsy 2023)
- Role of matrix proteins (M) and cellular cofactors in the assembly of influenza virus and SARS-CoV-2.
(Kerviel 2016; Bracquemond & Muriaux, 2021; Gourdelier 2023; Swain 2023)
- Atomic Force Microscopy (AFM) studies of HIV-1, IAV, and SARS-CoV-2 assembly on single viral particles.
(Faivre-Moskalenko 2014; Bernaud 2015; Lyonnais 2021)
- Study of viral replication dynamics (translation/amplification) in cells and during development.
(Dufourt et al., Science 2022; Bellec et al., Nat Comm 2022; Bellec et al., bioRxiv 2023)
1. Lipid sorting during the self-assembly of HIV-1 Gag and Influenza A M1
(Funding: ANR Fluobuds, CNRS, Univ Montp, GdR IMABIO, ERC Erasmus Mundus) This project aims to decipher the role of pre-existing or virus-induced lipid nanodomains in the host cell plasma membrane during viral assembly. In previous studies, we showed that the interaction between the matrix domain of the Gag protein and specific plasma membrane lipids (PS/PIP2) induces retrovirus assembly (Hamard-Péron et al., 2010; Hamard-Péron & Muriaux, 2011). We proposed that acidic lipid-enriched microdomains (ALEMs) are created by oligomerization of the HIV-1 Gag protein in the host cell plasma membrane during viral assembly (Kerviel et al., 2013, Yandrapalli et al., 2014, Mariani et al., 2014 & Saad & Muriaux, Editorial 2015). To address this, we characterized the nature and effects of the MA domain interaction of retroviral Gag proteins (HIV-1 and MLV) with lipid membranes in silico (Kerviel et al., 2013, Charlier et al., 2014). In vitro, we investigated the impact of HIV-1 Gag self-assembly on reorganization of the plasma membrane using model membranes whose molecular composition mimics that of the cellular membrane. We were able to show that during self-assembly, the viral Gag protein sorts membrane lipids and generates lipid nanodomains enriched in PIP2 and cholesterol while excluding sphingomyelin (Yandrapalli et al., 2016).
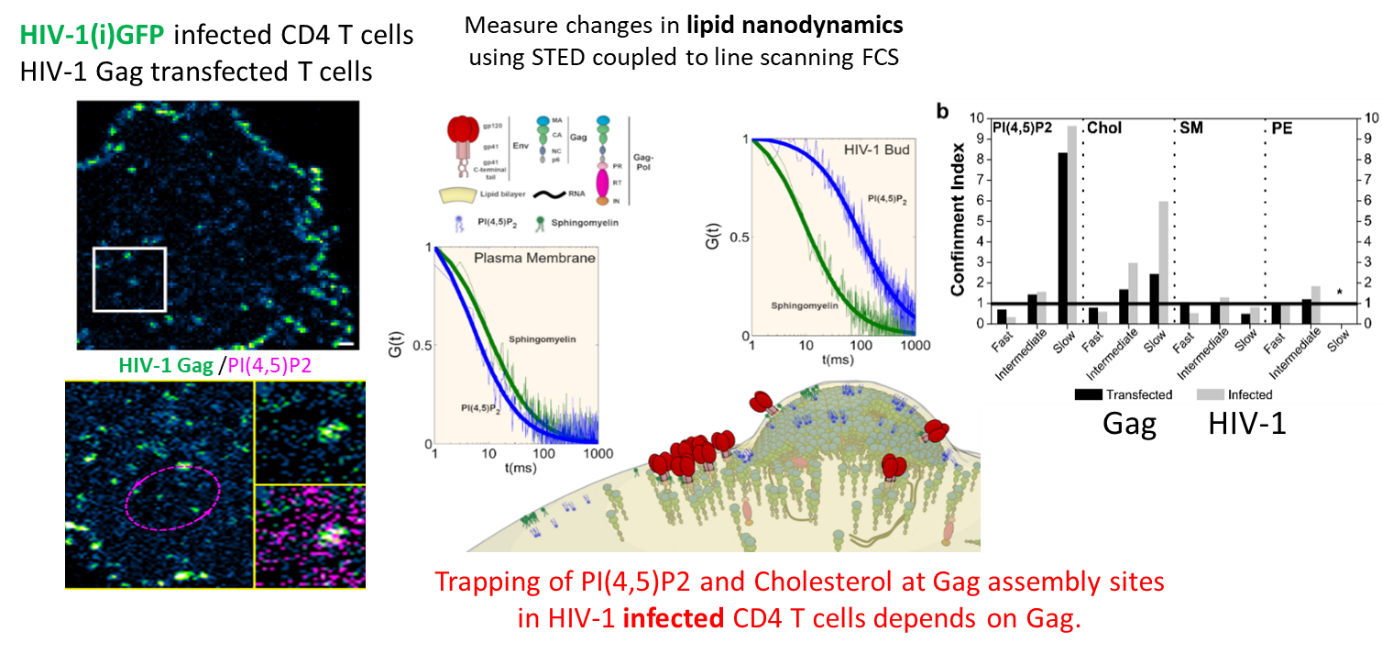 We finally quantified this phenomenon in the virus host cells (CD4 T lymphocytes infected with HIV-1 or transfected with Gag) by measuring changes in lipid diffusion (PIP2, cholesterol, SM and PE) inside and outside the viral bud at the surface of T cells using sSTED-FCS (Scanning STED FCS), in collaboration with C. Eggeling (U. Oxford/ Univ Jena, UK–Germany) and J. Chojnacki (U. Oxford/ IRSI Caixa, Barcelona, SP) (Favard et al., 2019).
We finally quantified this phenomenon in the virus host cells (CD4 T lymphocytes infected with HIV-1 or transfected with Gag) by measuring changes in lipid diffusion (PIP2, cholesterol, SM and PE) inside and outside the viral bud at the surface of T cells using sSTED-FCS (Scanning STED FCS), in collaboration with C. Eggeling (U. Oxford/ Univ Jena, UK–Germany) and J. Chojnacki (U. Oxford/ IRSI Caixa, Barcelona, SP) (Favard et al., 2019).
This work was highlighted by the Company of Biologists, by the INSB of the CNRS in its news and in its 2019 highlights, and by ANRS.
2. Single-molecule microscopy of HIV-1 assembly in living CD4 T lymphocytes
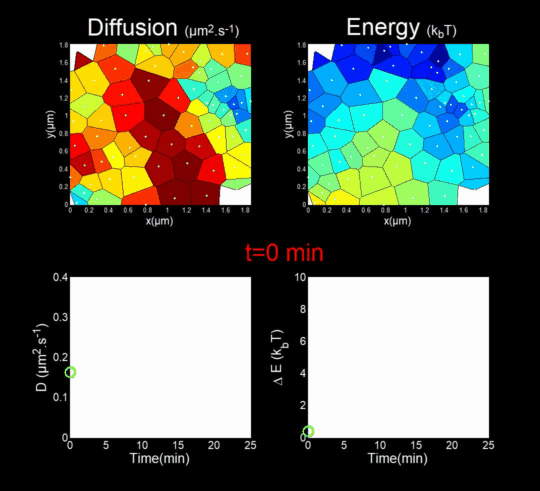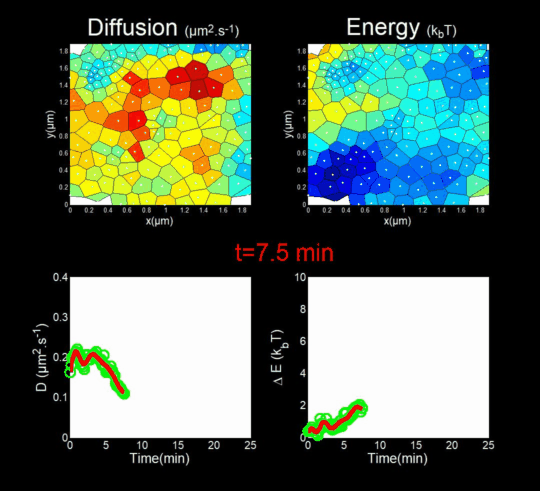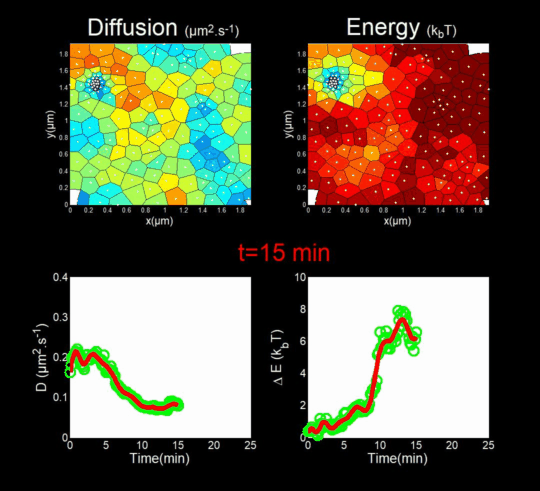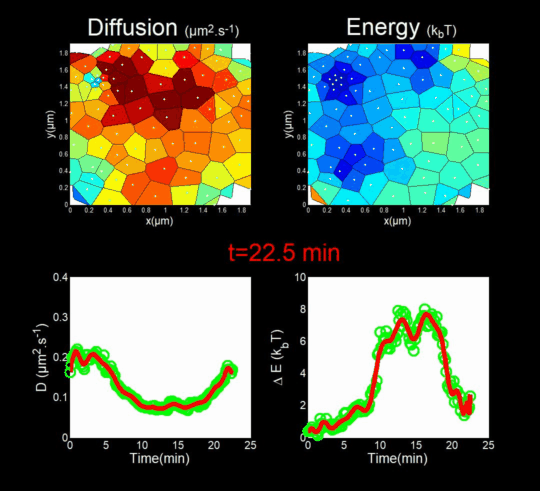
(Funding: ANR Fluobuds, CNRS, Univ Montp, GdR MIV, Erasmus Mundus)
In parallel, we study the dynamics of capsid protein assembly in cells using optical super-resolution microscopy techniques (Live PALM, spt-PALM), in collaboration with the teams of JB Sibarita (IINS, Bordeaux) and M. Dahan (Institut Curie, Paris).
We trace the assembly, molecule by molecule, with nanometer resolution, generating billions of individual trajectories (Mariani et al., 2016, Journal Cover).
We rely on Big Data methods for the automated analysis of these trajectories using Bayesian inference. In collaboration with J.B. Masson (Institut Pasteur), M. El Beheiry and M. Dahan (I. Curie), we were able to follow, over 25 minutes and molecule by molecule, the self-assembly of Gag during the formation of an HIV-1 VLP in a T lymphocyte. This included measuring both changes in molecular diffusion (left of the film) and the attractive energy (right of the film) experienced near the assembly site. (Mariani et al., 2018)
Our work on the study of HIV-1 assembly by live-cell super-resolution microscopy has led to two general-interest reviews (Inamdar et al., 2019 and Arone et al., 2021).
3. Cortical actin dynamics and membrane curvature during viral assembly
This project aims to identify host-cell cofactors from HIV-1 host CD4+ T cells that can jointly modulate dynamics, membrane domain organization, and membrane curvature during HIV-1 assembly, as well as filamentous cortical actin (F-actin).
In our research, we showed that the tetraspanin CD81 is a host cell transmembrane protein involved in HIV-1 assembly in CD4 T cells
(Grigorov et al., 2009) and that the clathrin–dynamin endocytic pathway is involved in HIV-1 transmission between CD4 T cells
(Bosch et al., 2008).
Our work now focuses on studying the role of these tetraspanins in the assembly of another human retrovirus, HTLV-1, which chronically infects T lymphocytes, in collaboration with Hélène Dutartre (CIRI, ENS Lyon) (funding ANRS 2021).
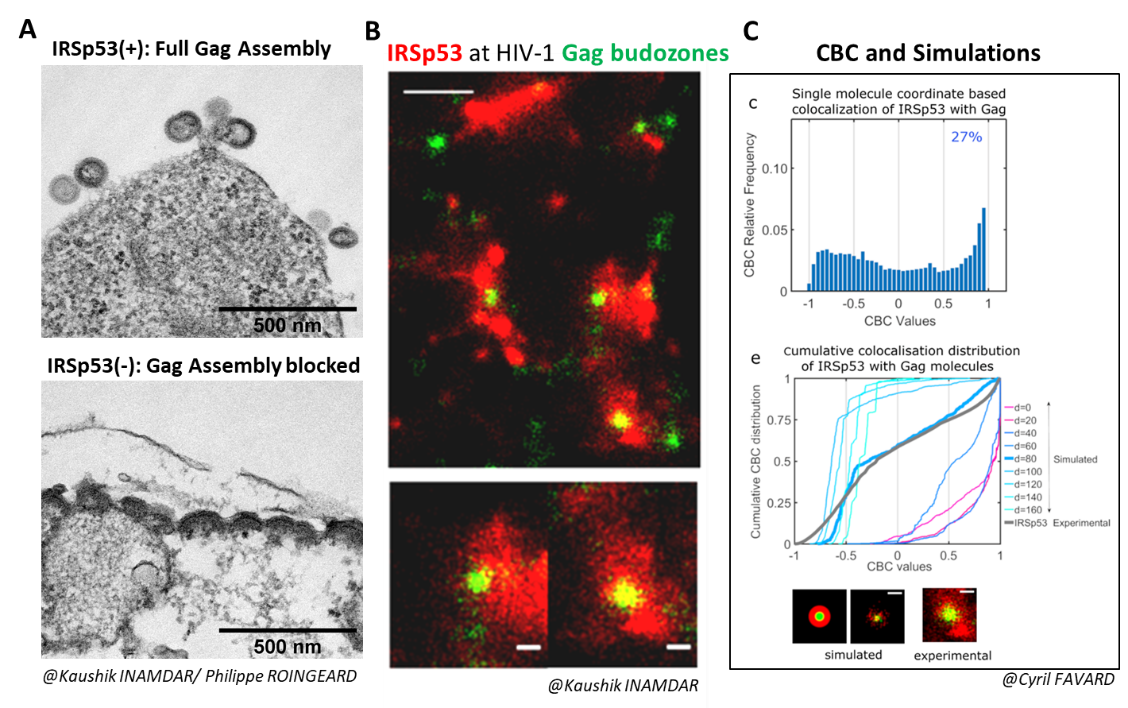
From Inamdar et al., 2021.
We also studied the role of a major regulator of the cortical actin network controlled by the Rac1/IRSp53/Wave2/arp2/3 activation pathway in the assembly of the HIV-1 Gag protein and the release of viral particles in CD4 T cells, the main hosts of HIV-1 (Thomas et al., 2015).
We demonstrated that the membrane curvature factor IRSp53 is involved in the assembly and budding of HIV-1 Gag viral particles (Inamdar et al., 2021).
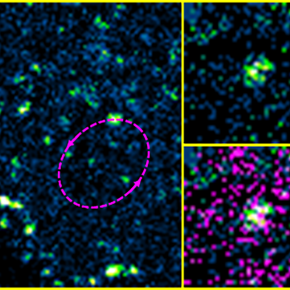
When IRSp53 expression was reduced by RNA interference in HIV-1 Gag-producing cells, we observed a mid-stage assembly block (Figure A).
IRSp53 was localized at the viral assembly site, as observed by PALM (Gag)/STORM (IRSp53) microscopy (Figure B), and quantified by correlating experimental single-molecule coordinate-based colocalization (CBC) measurements with numerical simulations on pre-established models (Figure C).
The cellular protein IRSp53 assists in membrane curvature during HIV-1 particle formation, enabling completion of viral assembly and budding.
Adapted from Inamdar et al., 2021.
4. Role of Matrix Proteins M and Cellular Cofactors in the Assembly of Influenza Virus and SARS-CoV-2
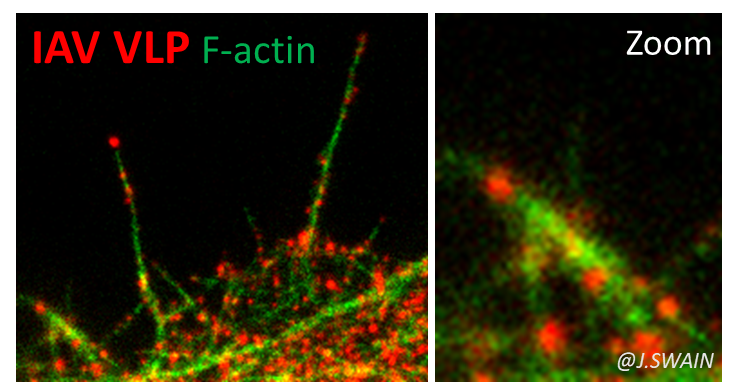
We study other enveloped RNA viruses, such as the influenza A (H1N1) pdm09 virus—still circulating and responsible for the first pandemic of the 21st century.
Our work focuses on the role of the matrix proteins M1 (and M2) in the membrane assembly of this virus. M1 is the most abundant viral protein and is essential for the assembly of the influenza virus.
We recently demonstrated that the arginine triplet (R76/77/78) located on helix 5 of M1—a highly conserved motif among influenza A subtypes—is indispensable for its localization to the plasma membrane, membrane binding of M1, its incorporation into viral particles, and viral infectivity
(in collaboration with O. Moncorgé, IRIM CNRS Montpellier, and P. Roingeard, University of Tours). Its mutation completely abolishes the production of infectious virus. This study also allowed us to establish a minimal system for producing non-infectious influenza A M VLPs (Kerviel, Dash et al., 2016).
We are now investigating the role of cortical actin and its regulators in the dynamics of assembly and budding of influenza A and SARS-CoV-2 viruses (Bracquemond & Muriaux, 2021),
using RNA interference techniques, infection assays, and advanced photonic microscopy coupled with cell biology.
5. Study by Atomic Force Microscopy (AFM) of HIV-1, IAV, and SARS-CoV-2 Assembly on Single Viral Particles
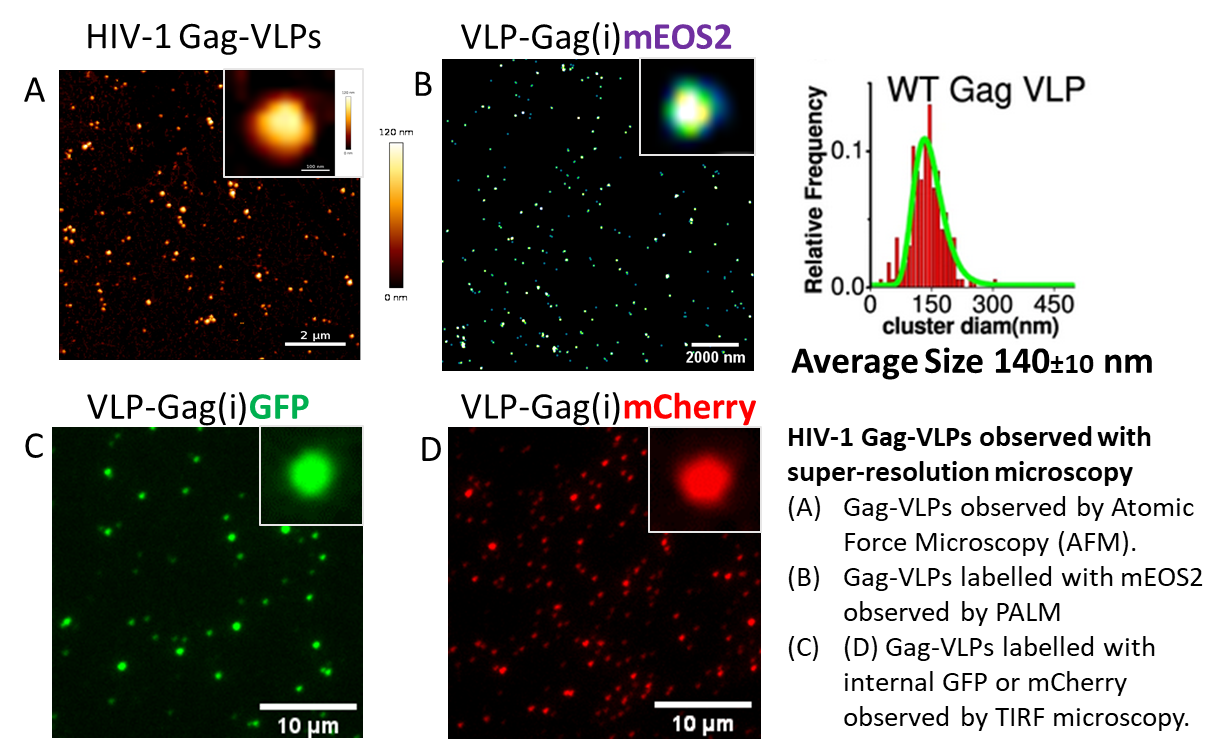
(Funding: ANRS 2015, ANRS 2019, MUSE)
Using atomic force microscopy at the level of single viral particles, we showed that the genomic RNA is a determinant of HIV-1 morphology polydispersity (Faivre-Moskalenko et al., 2014).
In collaboration with biophysicists C. Moskalenko and M. Castelnovo at ENS Lyon, AFM is used to quantitatively study the variation in size of viral particles and cores produced by host cells, depending on various microenvironments (Bernaud et al., 2015).
This polydispersity serves as an indicator of the efficiency of HIV-1 Gag protein self-assembly.We have recently imaged infectious and inactivated SARS-CoV-2 viruses by AFM, in collaboration with Sébastien Lyonnais and the Cemipai team (Lyonnais et al., 2021).
We continue to analyze various VLPs derived from HIV-1, IAV, and SARS-CoV-2 using fluorescence-based techniques, which provide powerful tools to study viral assembly and entry into host cells (outside of a BSL3 environment).




























 Rayane a brillament été sélectionnée pour présenter ses travaux sur le rôle de l’actine durant l’assemblage de HIV-1. De plus elle a obtenue une bourse de SIDACTION pour financer son déplacement au congrès.
Rayane a brillament été sélectionnée pour présenter ses travaux sur le rôle de l’actine durant l’assemblage de HIV-1. De plus elle a obtenue une bourse de SIDACTION pour financer son déplacement au congrès.